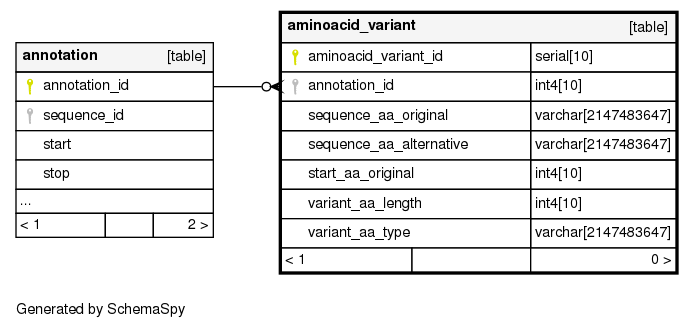
Columns
| Column | Type | Size | Nulls | Auto | Default | Children | Parents | Comments | ||
|---|---|---|---|---|---|---|---|---|---|---|
| aminoacid_variant_id | serial | 10 | √ | nextval('aminoacid_variant_aminoacid_variant_id_seq'::regclass) |
|
|||||
| annotation_id | int4 | 10 | null |
|
|
|||||
| sequence_aa_original | varchar | 2147483647 | null |
|
|
Affected amino acid sequence from the corresponding reference sequence of the chosen Virus |
||||
| sequence_aa_alternative | varchar | 2147483647 | null |
|
|
Changed amino acid sequence (in the target sequence) with respect to the reference one |
||||
| start_aa_original | int4 | 10 | √ | null |
|
|
Coordinate where the change starts on the reference sequrence |
|||
| variant_aa_length | int4 | 10 | null |
|
|
Length of amino acid change in units of impacted residues |
||||
| variant_aa_type | varchar | 2147483647 | null |
|
|
Type of amino acid change (SUB = substitution, INS = insertion, DEL = deletion) |
Indexes
| Constraint Name | Type | Sort | Column(s) |
|---|---|---|---|
| aminoacid_variant_pkey | Primary key | Asc | aminoacid_variant_id |
| aa__ann_id | Performance | Asc | annotation_id |
| aa__start_original | Performance | Asc | start_aa_original |
| aa__var_type_lower | Performance | ||
| aa__var_type_normal | Performance | Asc | variant_aa_type |