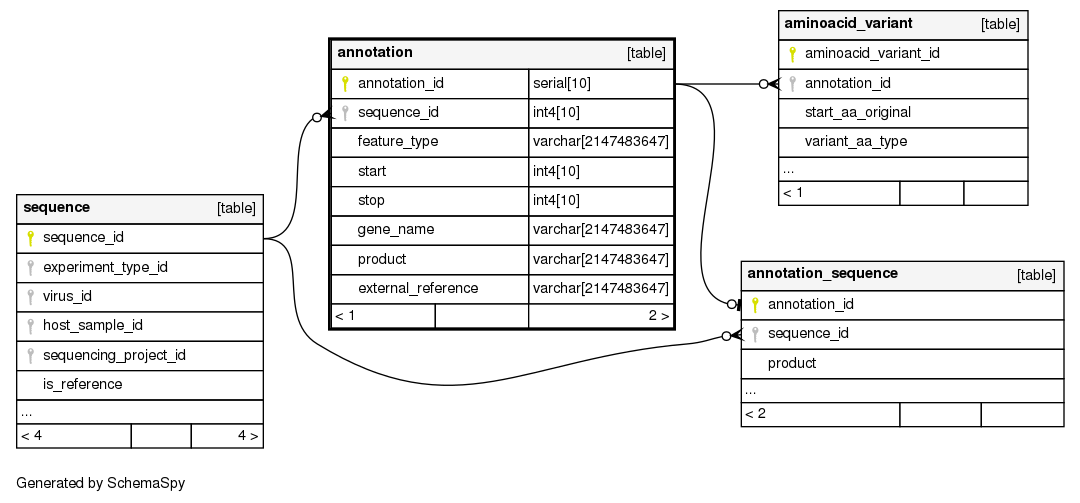
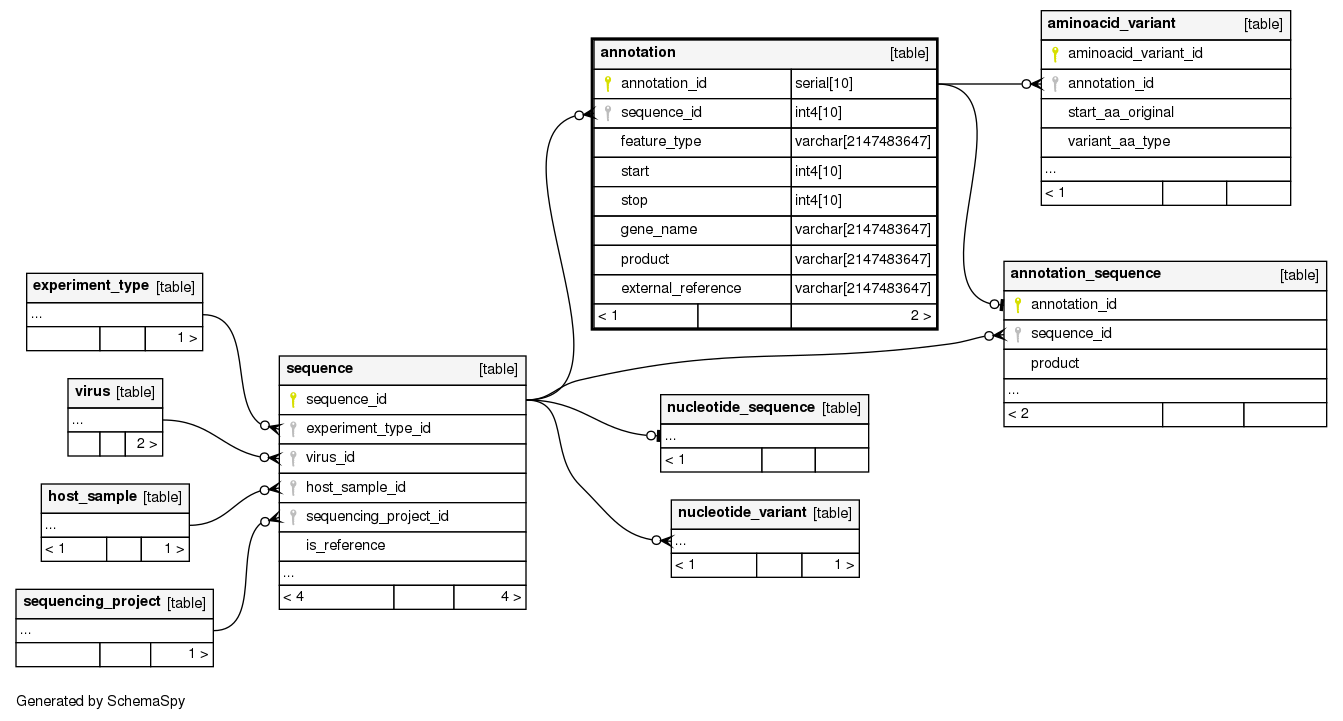
Columns
| Column | Type | Size | Nulls | Auto | Default | Children | Parents | Comments | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| annotation_id | serial | 10 | √ | nextval('annotation_annotation_id_seq'::regclass) |
|
|
||||||
| sequence_id | int4 | 10 | null |
|
|
|||||||
| feature_type | varchar | 2147483647 | null |
|
|
The annotation type of the observed sub-sequence (e.g., gene, peptide, coding DNA region, or untranslated region, molecule patterns such as stem loops and so on) |
||||||
| start | int4 | 10 | √ | null |
|
|
Start coordinate of annotation on reference sequence |
|||||
| stop | int4 | 10 | √ | null |
|
|
Stop coordinate of annotation on reference sequence |
|||||
| gene_name | varchar | 2147483647 | √ | null |
|
|
Gene to which the sub-sequence belongs |
|||||
| product | varchar | 2147483647 | √ | null |
|
|
Protein produced by the sub-sequence within which the amino acid change occurs |
|||||
| external_reference | varchar | 2147483647 | √ | null |
|
|
Eventually related external reference (when the protein is present in a separate database such as UniProtKB) |
Indexes
| Constraint Name | Type | Sort | Column(s) |
|---|---|---|---|
| annotation_pkey | Primary key | Asc | annotation_id |
| ann__seq_id | Performance | Asc | sequence_id |
| ann__start | Performance | Asc | start |
| ann__stop | Performance | Asc | stop |