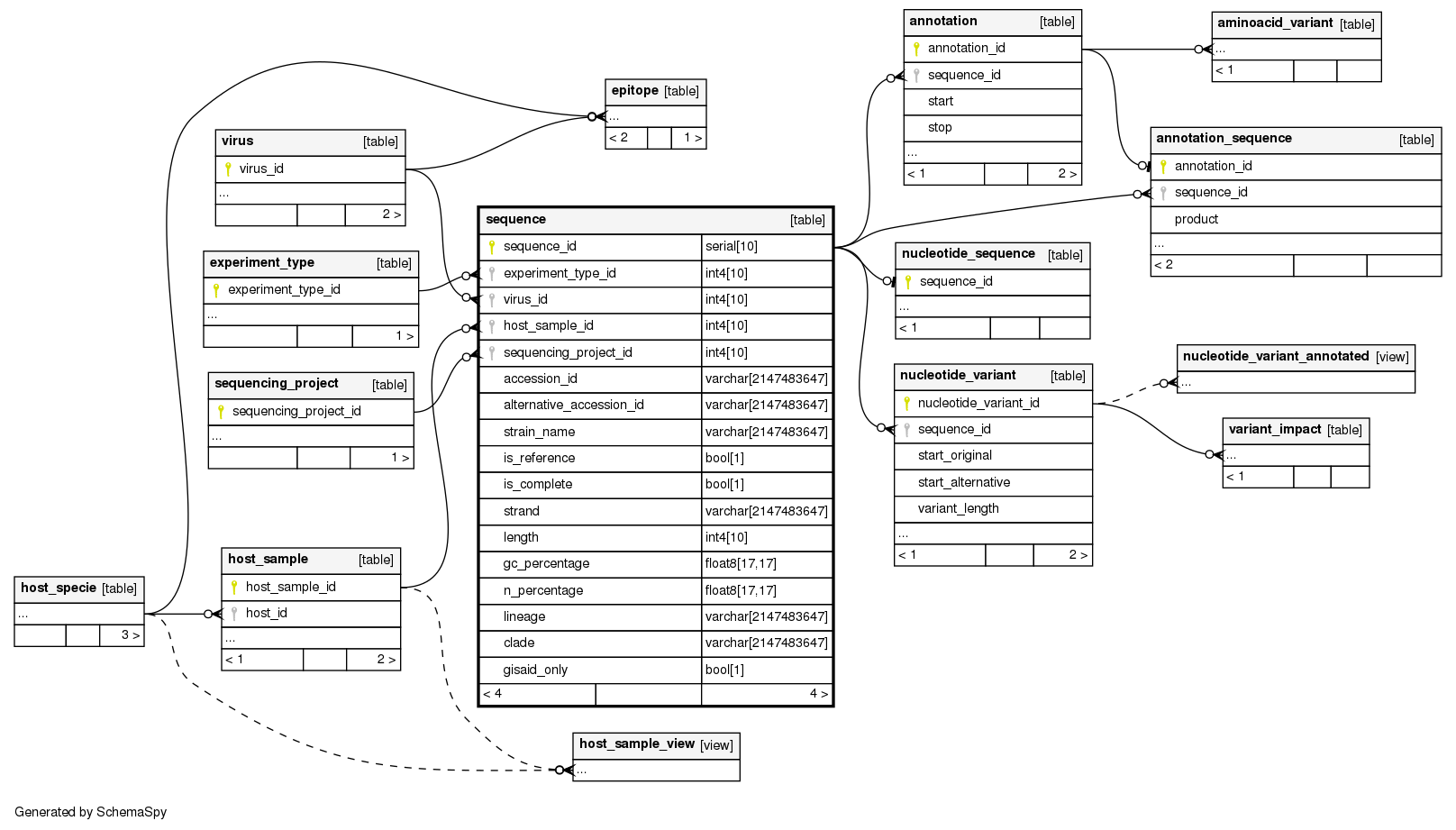
Columns
| Column | Type | Size | Nulls | Auto | Default | Children | Parents | Comments | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sequence_id | serial | 10 | √ | nextval('sequence_sequence_id_seq'::regclass) |
|
|
||||||||||
| experiment_type_id | int4 | 10 | null |
|
|
|||||||||||
| virus_id | int4 | 10 | null |
|
|
|||||||||||
| host_sample_id | int4 | 10 | null |
|
|
|||||||||||
| sequencing_project_id | int4 | 10 | null |
|
|
|||||||||||
| accession_id | varchar | 2147483647 | null |
|
|
Sequence identifier as extracted from the data source |
||||||||||
| alternative_accession_id | varchar | 2147483647 | √ | null |
|
|
Sequence alternative identifier as extracted from the original data source or another one |
|||||||||
| strain_name | varchar | 2147483647 | √ | null |
|
|
Name of strain of the sequence |
|||||||||
| is_reference | bool | 1 | null |
|
|
True when the sequence is the reference one (from RefSeq) for the virus species, False when the sequence is not the reference one |
||||||||||
| is_complete | bool | 1 | √ | null |
|
|
True when the sequence is complete, False when the sequence is partial. When not available from original source, we set False if its length is less than 95% of the reference sequence length, otherwise we set N/D since completeness cannot be determined with needed accuracy. |
|||||||||
| strand | varchar | 2147483647 | √ | null |
|
|
Strand to which the sequence belongs to (either positive or negative) |
|||||||||
| length | int4 | 10 | √ | null |
|
|
Number of nucleotides of the sequence |
|||||||||
| gc_percentage | float8 | 17,17 | √ | null |
|
|
Percentage of read G and C bases |
|||||||||
| n_percentage | float8 | 17,17 | √ | null |
|
|
Percentage of unknown bases |
|||||||||
| lineage | varchar | 2147483647 | √ | null |
|
|
Sequence lineage derived from source (for COG-UK) or calculated with the Pangolin software https://cov-lineages.org/pangolin.html (for other sources) |
|||||||||
| clade | varchar | 2147483647 | √ | null |
|
|
Clade as computed by GISAID (when available) |
|||||||||
| gisaid_only | bool | 1 | null |
|
|
True if sequence is only available via GISAID, False if available also in GenBank or COG-UK |
Indexes
| Constraint Name | Type | Sort | Column(s) |
|---|---|---|---|
| sequence_pkey | Primary key | Asc | sequence_id |
| seq__accession_id | Must be unique | ||
| seq__alternative_accession_id | Must be unique | ||
| seq__experiment_id | Performance | Asc | experiment_type_id |
| seq__host_id | Performance | Asc | host_sample_id |
| seq__seq_proj_id | Performance | Asc | sequencing_project_id |
| seq__virus_id | Performance | Asc | virus_id |
| sequence__is_reference__idx | Performance | Asc | is_reference |