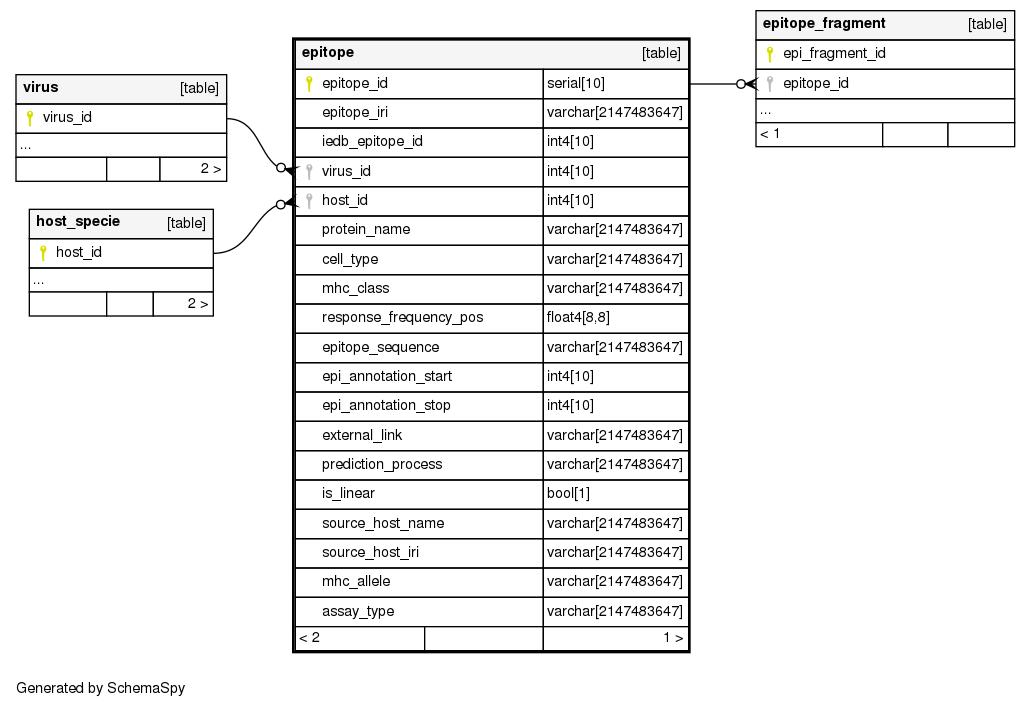
Columns
| Column | Type | Size | Nulls | Auto | Default | Children | Parents | Comments | ||
|---|---|---|---|---|---|---|---|---|---|---|
| epitope_id | serial | 10 | √ | nextval('epitope_epitope_id_seq'::regclass) |
|
|||||
| epitope_iri | varchar | 2147483647 | √ | null |
|
|
Identifies the single epitope within the IEDB system (ID) and allows to resort to its information |
|||
| iedb_epitope_id | int4 | 10 | √ | null |
|
|
Identifies the single epitope within the IEDB system (IRI) and allows to resort to its information |
|||
| virus_id | int4 | 10 | null |
|
|
|||||
| host_id | int4 | 10 | null |
|
|
|||||
| protein_name | varchar | 2147483647 | √ | null |
|
|
||||
| cell_type | varchar | 2147483647 | √ | null |
|
|
Indicates the target of the experiment (allowing values are ‘T cell’, ‘B cell’, ‘MHC ligand’) |
|||
| mhc_class | varchar | 2147483647 | √ | null |
|
|
Indicates the general classes of alleles provided in the previous field (possible values are ‘I’, ‘II’, or ‘I,II’ if both class I and II alleles are considered) |
|||
| response_frequency_pos | float4 | 8,8 | √ | null |
|
|
On IEDB this measure is defined as the number of positively responded subjects (R) divided by the total number of those tested (N), summed up by mapped epitopes; however, to compensate for epitopes that are identified by a low number of assays, we employ a corrected formula (proposed in Carrasco et al., 2015), where the importance of corrections decreases as the number of assays increases. |
|||
| epitope_sequence | varchar | 2147483647 | √ | null |
|
|
Indicates the general classes of alleles provided in the previous field (possible values are ‘I’, ‘II’, or ‘I,II’ if both class I and II alleles are considered). |
|||
| epi_annotation_start | int4 | 10 | √ | null |
|
|
Start coordinate of epitope on protein |
|||
| epi_annotation_stop | int4 | 10 | √ | null |
|
|
Stop coordinate of epitope on protein |
|||
| external_link | varchar | 2147483647 | √ | null |
|
|
References the publication where the epitope was first reported (and manually collected by IEDB curation team). |
|||
| prediction_process | varchar | 2147483647 | √ | null |
|
|
Prediction process used for the epitope (now they are all imported from IEDB) |
|||
| is_linear | bool | 1 | √ | null |
|
|
Defines continuous (true) or discontinuous (false) epitopes, composed of amino acid residues that may be located on different protein regions – brought together by protein folding |
|||
| source_host_name | varchar | 2147483647 | √ | null |
|
|
||||
| source_host_iri | varchar | 2147483647 | √ | null |
|
|
||||
| mhc_allele | varchar | 2147483647 | √ | null |
|
|
Also referred to as ‘mhc_allele’, it indicates the list of the class (e.g., ‘HLA Class I’), or lists of alleles (e.g., ‘HLA-B*35:01, HLA-B*15:01’) to which the epitope is restricted-this is relevant only for T-cell and MHC Ligand assays |
|||
| assay_type | varchar | 2147483647 | √ | null |
|
|
Indicates the outcome - considering possibly multiple experiments (we have ‘positive’, ‘negative’, and ‘both’, when positive and negative outcomes were included) |
Indexes
| Constraint Name | Type | Sort | Column(s) |
|---|---|---|---|
| epitope_pkey | Primary key | Asc | epitope_id |